Tumor classification using molecular signatures
Classifying malignant tumors has typically relied on pathologic criteria from the tissue site of origin with histologic and other clinical characteristics of the tumor determining the target and type of therapeutic intervention. This approach to classifying cancer, however, is slowly being rethought.
Classifying malignant tumors has typically relied on pathologic criteria from the tissue site of origin with histologic and other clinical characteristics of the tumor determining the target and type of therapeutic intervention.1 This approach to classifying cancer, however, is slowly being rethought as a more precise understanding of the molecular characteristics of tumors is emerging from several large-scale genomics projects.
Two primary observations emerging from the molecular analysis of cancer are that 1) cancers from the same organ are often distinct from each other, and 2) cancers of different organs share many of the same features.2,3 These emerging insights are paving the way for the potential to better target the specific pathologic pathways underlying disease and build a stronger bridge toward personalized medicine in oncology.
This article briefly describes the ongoing research and current findings that are reshaping the classification of cancer and leading, it is hoped, to better, more precise treatments. Focus is on the extensive work being done by The Cancer Genome Atlas (TCGA) Research Network and TCGA Pan-Cancer Initiative, which are providing the foundation for the molecular analysis of tumors.2,3 The article also briefly describes genomic research under way in childhood cancers, highlighting one of several genomic initiatives over the past 5 years aimed at cataloging all the genomic lesions in childhood cancer.
TCGA Research Network and TCGA Pan-Cancer Initiative
In 2006, the TCGA Research Network project was initiated with a goal of collecting and profiling tumor samples from at least 20 tumor types to discover molecular aberrations at the genetic and epigenetic levels.1,2 To date, the TCGA Research Network has identified 12 primary tumor types (Table 1).3

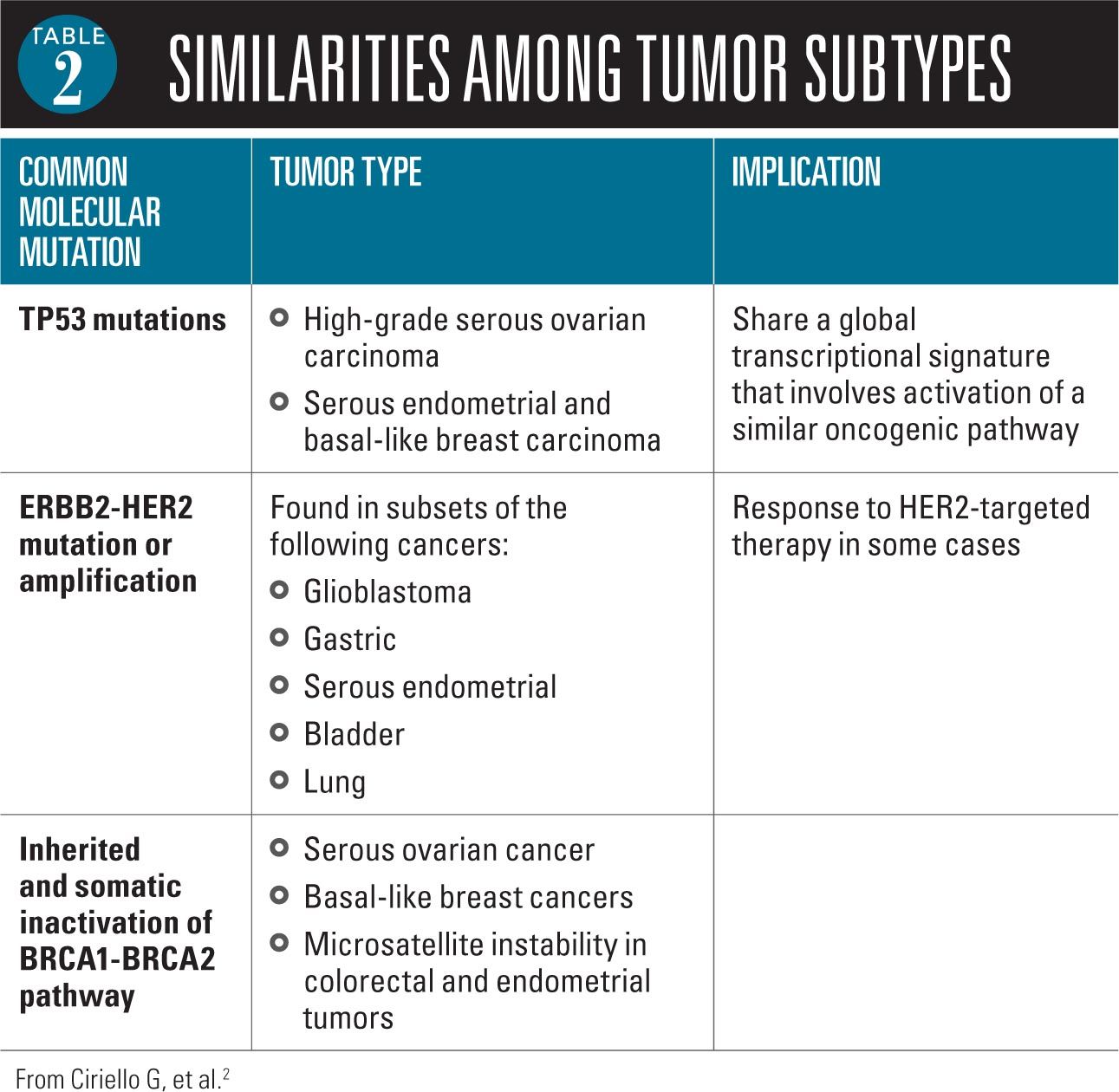
Two primary observations emerged from the molecular analysis of cancer: 1) cancers from the same organ are often distinct from each other, and 2) cancers of different organs share many of the same features.2 Researchers have identified, for example, a number of important similarities among tumor subtypes from different organs (Table 2).
To help coordinate and provide a systematic foundation from which to discover differences, commonalities, and emergent themes across tumor lineages, the TCGA Pan-Cancer Initiative was launched in 2012 with the specific aim of comparing the first 12 tumor types identified through the TCGA Research Network.3 The initiative involves over 250 collaborators from 30 institutions using the same data set to work on over 60 different research projects.4
In 2013, the first findings of this initiative were published.2,5 A study by Ciriello and colleagues found that tumors comprise 2 major genomic categories independent of tumor tissue type or tissue of origin. Tumors had either a large number of copy number alterations or they had a large number of somatic mutations.2 Furthermore, the study found that tumors across several tissue types share the same oncogenic signature. Based on these findings, the investigators derived a hierarchical classification in terms of oncologenic signatures of thousands of tumors from the 12 tumor types.
A second study by Zack and colleagues, also published in 2013, provides insight into the mechanisms of generation and functional consequences of cancer-related somatic copy number alterations (SCNAs), which play a critical role in activating oncogenes and inactivating tumor suppressors.5 The investigators identified common patterns of SCNAs across cancer type and found significant recurrent focal SCNAs in 140 genomic regions. Among these regions, 102 did not have known oncogene or tumor suppressor gene targets and 50 regions had a significant number of mutated genes.
In a commentary accompanying the studies, John Weinstein, MD, PhD, University of Texas MD Anderson Cancer Center, Houston, and colleagues said that “shared molecular patterns will enable etiologic and therapeutic discoveries in one disease that can be applied to another. Importantly, integrative interpretation of the data will help identify how the consequences of mutations vary across tissues, with important therapeutic implications.”3
“Relatively rare cancers, such as childhood malignancies, in particular stand to benefit from such an approach,“ the researchers said in the commentary.3
In the most recent study published in 2014, Hoadley and colleagues further examined the molecular alterations of the 12 tumor types to see which alterations are shared across cancers arising from different tissues.1 They also looked at whether disease subtypes previously identified do span multiple tissues of origin.
To test their hypothesis that molecular signatures provide a distinct molecular taxonomy relative to the current classification of tumors by tissue of origin, the investigators used a multiplatform integrative analysis of thousands of cancers from the 12 tumor types.1
Using the data from multiple assay platforms, the investigators identified 11 major subtypes.1 Most subtypes were identified by tissue of origin features, but several distinct cancer types converged into common subtypes. One subtype typified by TP53 alterations, TP63 amplifications, and high expression of immune and proliferation pathway genes included lung squamous cancers, head and neck cancers, and a subset of bladder cancers. The most heterogeneous malignancy was bladder cancers that split into 3 primary subtypes.
Overall, the study found that about 10% of cases were reclassified by the molecular taxonomy, indicating that 1 in 10 patients would be classified differently by the new molecular taxonomy versus using just the current tissue of origin system of classifying tumors.1
The implication of these findings is the potential to better target therapy by improving the ability to more precisely subtype cancers. “If used to guide therapeutic decisions, this reclassification would affect a significant number of patients to be considered for nonstandard treatment regimens,” state the investigators. “In addition to identifying several new genomic and pathway insights between and within tissue-of-origin tumor types, this TCGA study provides a public resource compendium of individuals and integrated data sets . . . enabling researchers to explore new questions and analytical approaches that will perpetuate this discovery process.”1
Looking for molecular signatures for childhood cancers
Over the past 5 years, a number of large genomic initiatives have been started to catalog all the genomic lesions present in childhood cancer. Among these is a project through the National Cancer Institute (NCI) called Therapeutically Applicable Research to Generate Effective Treatments (TARGET) managed by the Office of Cancer Genomics and Cancer Therapy Evaluation Program. Genomic data generated from the TARGET initiative is available to the research community with the broad aim of facilitating the discovery of therapeutic targets for childhood cancers and translating this into clinical application. Current projects include research that is examining the genomes, transcriptions, and/or epigenomes of selected childhood cancers including acute lymphoblastic leukemia, acute myeloid leukemia, kidney tumors, neuroblastoma, and osteosarcoma.
According to Malcolm A. Smith, MD, PhD, associate branch chief, Pediatrics, in the Clinical Investigations Branch, Cancer Therapy Evaluation Program, Division of Cancer Treatment and Diagnosis, NCI, Bethesda, Maryland, enormous progress has been made over the last 5 years as a result of the large-scale initiatives that together have sequenced thousands of childhood cancer genomes. One major insight that has emerged is the recognition that diseases previously thought to be relatively homogenous actually represent multiple molecularly distinctive subtypes. For example, he said, it is now recognized that there are 4 distinctive subgroups within medulloblastoma that have different demographic and prognostic characteristics, different genomic lesions, and different potentials for being treated with specific targeted therapies.
“We need to adjust our thinking about how we approach medulloblastoma diagnostically and eventually how we treat it,“ said Smith, adding that the information gathered from genomic research is being used in the development of medulloblastoma clinical trials. Similar molecularly defined subtypes have been identified for other cancers and are being incorporated into clinical trials conducted by clinical trial groups such as the Children’s Oncology Group.
Saying that genomic information will be increasingly used over the next years, Smith emphasized that this information will serve to complement the information gleaned from histology and other clinical characteristics but is unlikely to replace tissue-of-origin classification of disease. “Tissue of origin is still important, and the genomic information will complement, extend, and enrich it to potentially point to specific treatments in some cases and in other cases to provide prognostic information that can help guide treatment,” he said.
Building the bridge to personalized medicine
All this research points to finding a better way to tailor treatment and provide the best care possible to people with cancer. “The hope is that investigations across tumor type such as the Pan-Cancer project will ultimately inform clinical decision making,” according to Weinstein and colleagues.3 “We hope [such studies] will enable discovery of novel therapeutic agents that can be tested clinically-perhaps in novel adaptive, biomarker-based clinical trials that cross tumour boundaries.”
Success in using genomic information already has been seen with the development of recent cancer treatments that specifically target molecular changes now recognized in specific cancers.6 These targeted therapies include imatinib that inhibits an altered enzyme found in patients with chronic myelogenous leukemia; trastuzumab that targets human epidermal growth factor receptor 2 (HER-2) mutations in patients with HER-2 positive breast cancer; and gefitinib and erlotinib that target epidermal growth factor receptor (EGFR) mutation in patients with EGFR positive lung cancer.
In addition, genomics research provides a better understanding of which patients will not benefit from selected therapies, and thereby helps to avoid unnecessary treatment and adverse effects.6 This can be seen with cetuximab and panitumumab, both targeted therapies that do not benefit colon cancer patients with tumors that have a mutation in a gene called KRAS.
The broad aim of collaborative efforts by initiatives such as the Pan-Cancer project and TARGET is to increasingly expand the molecular analysis of cancers to more tumor types with the long goal of developing more target-specific treatments and, ultimately, offering patients more precise personalized medicine.
REFERENCES
1. Hoadley KA, Yau C, Wolf DM, et al; Cancer Genome Atlas Network. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929-944.
2. Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45(10):1127-1133.
3. Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, et al. The Cancer Genome Atlas Pan-Cancer analysis project. Nat Genet. 2013;45(10):1113-1120.
4. Omberg L, Ellrott K, Yuan Y, et al. Enabling transparent and collaborative computational analysis of 12 tumor types within the Cancer Genome Atlas. Nat Genet. 2013;45(10):1121-1126.
5. Zack TI, Schumacher SE, Carter SL, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45(10):1134-1140.
6. The Cancer Genome Atlas (TCGA). Impact of cancer genomics on precision medicine for the treatment of cancer. TCGA website. Available at: http://cancergenome.nih.gov/cancergenomics/impact. Accessed August 26, 2012.
Ms Nierengarten, a medical writer in St. Paul, Minnesota, has over 25 years of medical writing experience, coauthoring articles for Lancet Oncology, Lancet Neurology, Lancet Infectious Diseases, and Medscape. The author has nothing to disclose in regard to affiliations with or financial interests in any organizations that may have an interest in any part of this article.