Genetic testing for intellectual disability: A role in diagnostic evaluation
Intellectual disability (ID) is a condition with a prevalence of 1% to 3%. As a result, most pediatricians will be required to address and manage this condition on a regular basis.
Intellectual disability (ID) is a condition with a prevalence of 1% to 3%.1-5 As a result, most pediatricians will be required to address and manage this condition on a regular basis. In many circumstances an underlying etiology has been identified, but often the diagnosis of ID or global developmental delay (GDD) is made by a pediatrician and there is no immediately apparent underlying explanation. Establishing an underlying diagnosis has numerous implications beyond just medical treatment and should be considered a priority of patient care.
There are numerous causes of ID, but prenatal causes are by far the most common. Of these, genetic abnormalities predominate. If a diagnosis is not made after conducting an appropriate history and physical examination, genetic testing, and specifically a chromosomal microarray, is considered the first-line procedure in the diagnostic evaluation of ID, according to the American Academy of Neurology, the Child Neurology Society, and the American College of Medical Genetics.1,4,6 This article reviews the role of chromosomal microarray in the diagnosis of ID/GDD.
Definition of ID/GDD and importance of a diagnosis
Intellectual disability is defined as consistently subaverage intellectual function that is accompanied by defects in adaptive, conceptual, or social skills with onset before 18 years of age.3 The diagnosis and subclassification into mild, moderate, and severe ID are made based on IQ testing; an IQ of less than 70 is the minimum criterion for mild ID. GDD is defined as performance that is 2 standard deviations below age-appropriate norms in 2 or more areas of development and is a more useful definition in children who are aged younger than 6 years for whom IQ testing cannot be performed.1 GDD is often a precursor to ID, and pediatricians are obligated to conduct a diagnostic evaluation before a child is capable of completing IQ testing. Therefore, ID and GDD are used interchangeably here.
Establishing a diagnosis of ID can be labor intensive and may require the involvement of subspecialists. However, regardless of the availability of specific treatment or cure, knowing the diagnosis may have far-reaching benefits for the patient and family and justifies the effort. Identifying an underlying genetic cause may give a better indication of prognosis as well as recurrence risk for the patient and family. Along those same lines, a genetic diagnosis may provide the ability to anticipate associated comorbidities and other affected organ systems that have not yet been recognized. From a practical standpoint, a diagnosis will also help families gain access to services. These services may range from disease-specific therapies to financial assistance and support groups, all of which are likely to improve quality of life for both the patient and family and should not be undervalued.
Numerous causes of ID have been identified. These include, but are not limited to, genetic abnormalities, infection, trauma, complications of extreme prematurity, toxic exposures, hypoxia, hemorrhage, malnutrition, or metabolic and endocrine abnormalities. Of the metabolic, toxic, and infectious etiologies, prenatal and neonatal injuries are the most common. A child with ID may have one of these causes readily identifiable. However, up to 60% to 80% of children will not have a readily identifiable underlying diagnosis.7,8 A genetic evaluation is therefore indicated as part of the diagnostic evaluation of these children. Ideally, a consultant with genetics training would guide this process. However, time constraints may make this difficult, and geographic proximity would be necessary. An understanding of the utility of current diagnostic techniques may enable the pediatrician to expedite this process.
Clinical evaluation of ID/GDD
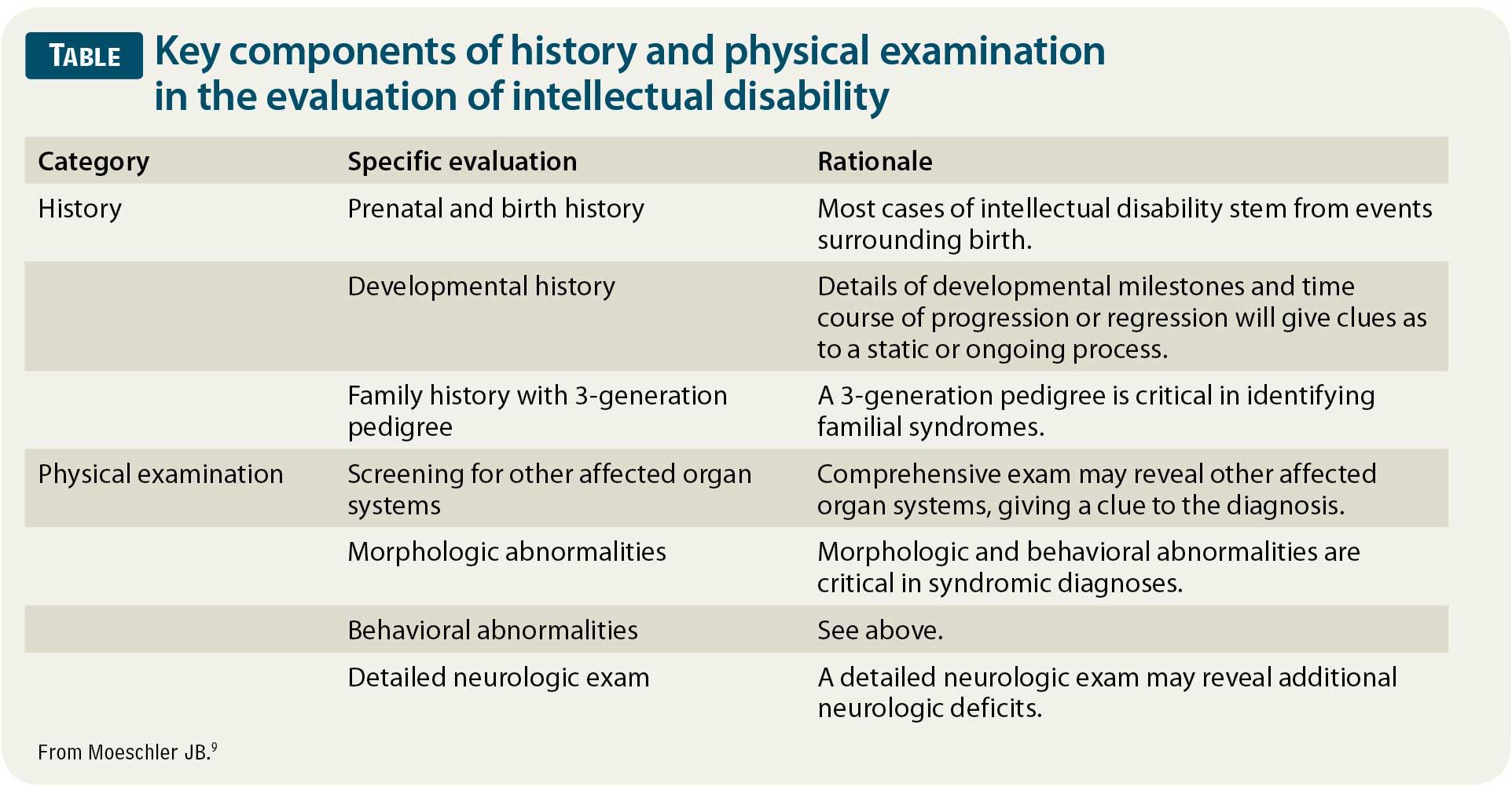
As with all pediatric illnesses and conditions, a detailed history and physical examination are paramount when evaluating the underlying etiology of ID (Table).9 Various studies have suggested that the history and physical examination alone are responsible for a diagnosis in one-third to two-thirds of cases of ID. The history should focus on prenatal and birth history, developmental history, and family history. Additionally, given the prevalence of genetic disorders in ID, a 3-generation pedigree should be obtained when possible. The physical examination should be comprehensive, with a high suspicion for other affected organ systems that may give a clue to the diagnosis. A detailed neurologic examination should be conducted to look for other signs of diffuse or focal abnormalities that may indicate a need for neuroimaging. These signs include abnormalities in strength, tone, and coordination, especially when any findings are asymmetrical. Finally, attention should be paid to any morphologic or behavioral abnormalities that may aid in narrowing the differential diagnosis given their association with specific syndromes. Some examples include macrocephaly or microcephaly; abnormal positioning of the eyes and ears; overdeveloped or underdeveloped sexual characteristics; and behaviors such as tics, abnormal movements, aggression, or inappropriate affection.
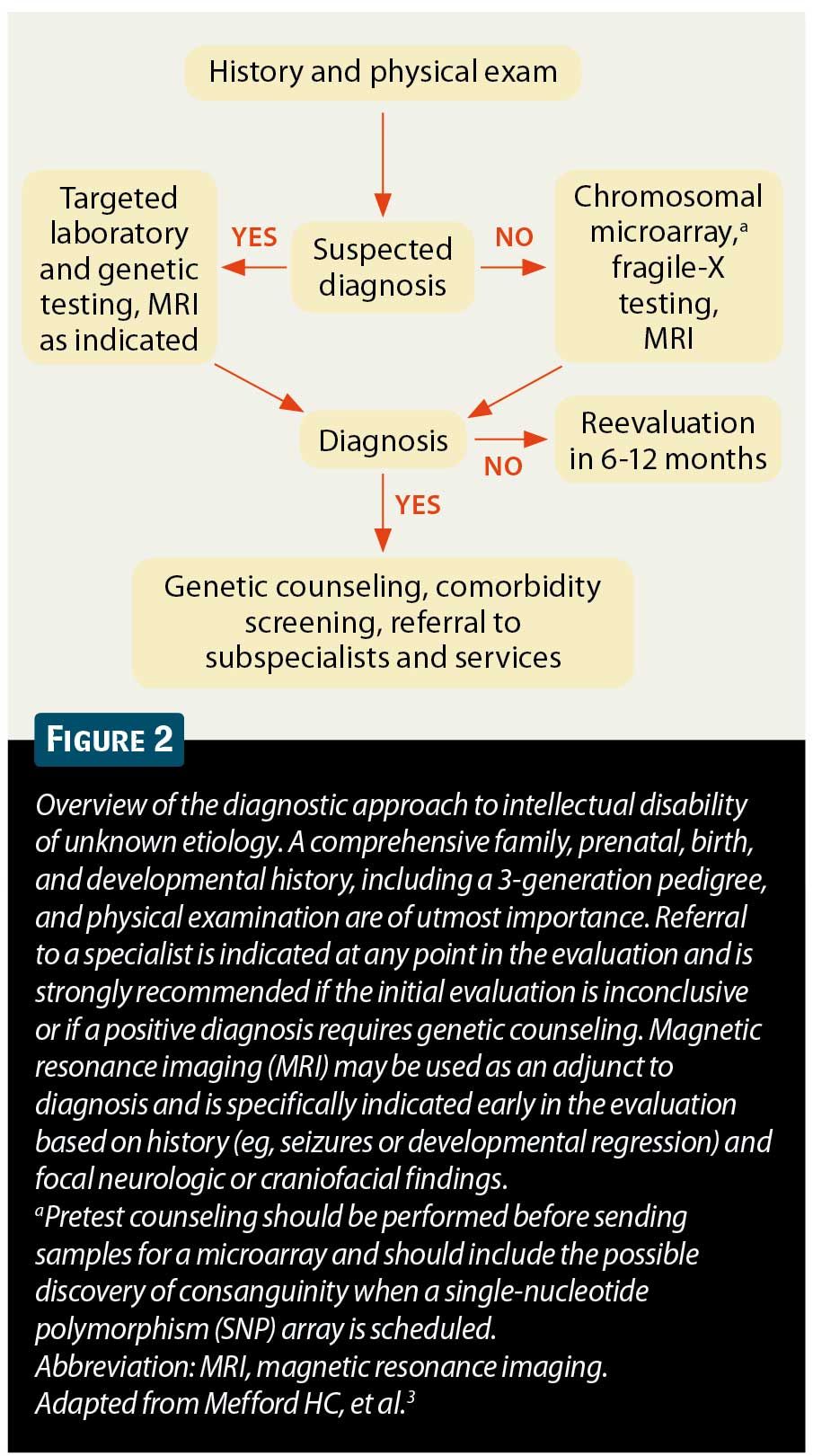
Upon completion of the history and physical examination, there are multiple possible results that will determine the type and extent of further testing. A diagnosis may be made based on history and physical examination alone (eg, the history reveals significant intrauterine alcohol exposure and the patient has morphology and behavior consistent with fetal alcohol syndrome). In this case, further testing may not be warranted. However, more commonly, the history and physical examination suggest etiologies that require further testing. A suspected metabolic or endocrine abnormality requires diagnostic or screening laboratory testing (eg, thyroid function studies for hypothyroidism or screening fasting plasma amino acids and urine organic acids for suspected inborn errors of metabolism). Similarly, a specific genetic syndrome may be suspected but requires confirmation (eg, specific testing for Rett syndrome in girls with developmental regression and microcephaly, or G-banded karyotype for infants with examination findings consistent with trisomy 21). Finally, patients may have isolated ID with or without focal neurologic findings but with no hints as to an underlying etiology. In these 2 groups of patients, genetic testing is critical. Chromosomal microarray along with fragile-X testing is currently considered the most sensitive and comprehensive test when a specific diagnosis is not suspected.1,4,6
Genetic testing for ID/GDD
Karyotyping was first used to identify trisomy 21 in 1959. Since that time, genetic testing has evolved tremendously, and the question of what modality is most clinically useful and cost-effective has become more pertinent. During the past 5 years, multiple studies have clearly demonstrated that chromosomal arrays are the highest-yielding diagnostic tools for ID of unknown etiology and are preferred over G-banded karyotypes (standard karyotyping) and fluorescence in situ hybridization (FISH) assays. The exception would be cases of clinically recognizable aneuploidy syndromes or a family history of balanced translocations or multiple spontaneous abortions.1,4,10-15
Chromosome microarrays are assays that use fluorescence-based technology to detect copy-number changes (duplications and deletions) across the genome. There are 2 commonly used types of arrays.6,10 The first is comparative genomic hybridization (CGH; Figure 1A). This technique compares the amount of fluorescently labeled DNA from a patient sample that is bound to known DNA sequences to the amount of DNA from a healthy control sample that binds to the same DNA sequences. This type is usually called an oligoarray and spans the length of all chromosomes, with enrichment in known areas of copy-number variation. Most oligoarrays have 144,000 to 180,000 probes. The second type of array is a single-nucleotide polymorphism (SNP) genotyping array (Figure 1B). SNP arrays take advantage of multiple sites in the genome where 2 different alleles are present in the general population. The 2 different alleles are differentially fluorescently labeled and hybridized with patient DNA. The total fluorescence and the fluorescence ratio of the 2 different dyes allow analysis of homozygosity and heterozygosity as well as identification of duplications or deletions. Most SNP arrays detect 660,000 to 2 million SNPs across the length of all chromosomes.
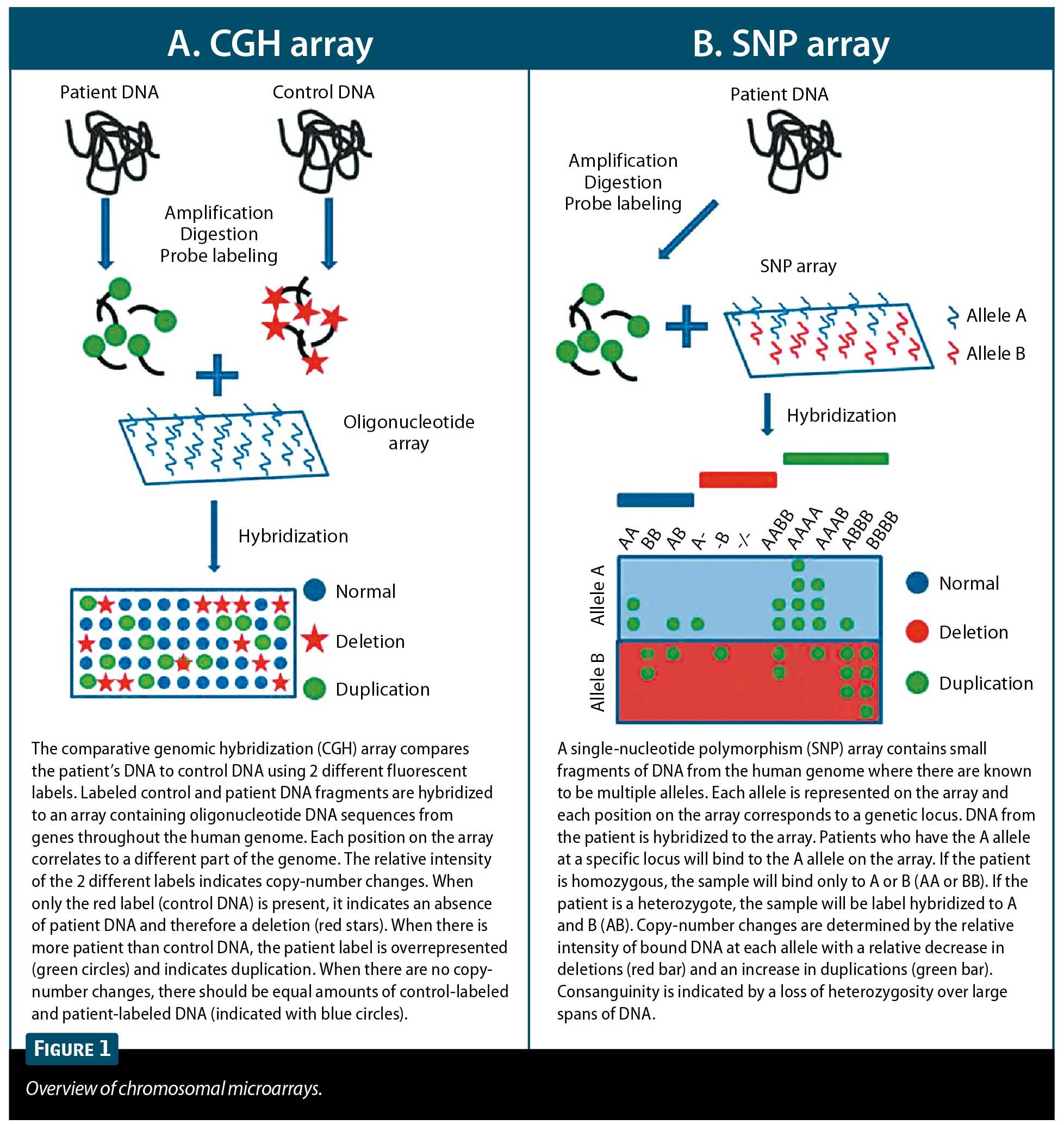
It should be noted that SNP arrays are capable of detecting consanguinity and uniparental disomy, whereas CGH arrays are not.3,10,16 Studies looking at the diagnostic yield of CGH and SNP arrays have reported a yield of 10% to 30%, with the majority of studies reporting 15% to 20%.1,4,10-15 In contrast, G-banded karyotyping detects abnormalities in only 2% to 4% of cases, and FISH in 2.4% to 3.5%.1,4
It has been argued that the use of microarrays will cause an inability to detect balanced translocations. Balanced translocations result from an exchange of material between nonhomologous chromosomes resulting in the same absolute amount of DNA that is located on a different chromosome. This creates the potential for interruption of coding or regulatory sequences. Because a microarray detects only copy-number variation (how many copies of a sequence are present) and not organization (where the sequence is present), there is a theoretical risk of missing a balanced translocation. However, many balanced translocations at the resolution of the G-banded karyotype are not balanced at the molecular level, and there is a copy-number change that can be detected by microarray. Additionally, balanced rearrangements account for only a tiny minority of changes in patients with ID, and the routine use of G-banded karyotyping is not indicated to capture these events. Further, chromosomal microarrays have a similar ability to detect mosaicism as G-banded karyotyping. Generally, 20 cells are analyzed in a routine karyotype, and a minimum of 3 abnormal cells is indicated before 50 cells are analyzed. Thus, the lower limit for detection of mosaicism is 14%, and most laboratories quote 20%. The CGH array is comparably sensitive. The SNP arrays can detect mosaicism at a resolution of 5%.4
Although the chromosomal array should be the standard testing modality in most cases, there are limited indications for the use of G-banded karyotype in addition to or instead of an array. These scenarios include clinically recognizable aneuploidy syndromes such as trisomy 21, 18, or 13, or Turner (45,X) or Klinefelter (47,XXY) syndromes; more than 2 spontaneous abortions; or a known family history of balanced translocations.4 In the latter 2 cases, there is likely to be a parental chromosomal abnormality present. Therefore, karyotyping of the parents would also be recommended for further characterization and genetic counseling.
Although trisomy 21 is the most common chromosomal abnormality associated with ID, fragile X syndrome is the most common single-gene defect linked to ID and accounts for 1% of all males and about 0.3% of females with ID. Although postpubertal males develop a long face and macroorchidism, younger boys and affected girls have no specific phenotype. Therefore, fragile-X testing should be performed in addition to a microarray in the initial evaluation of nonsyndromic ID.9
Some specific syndromes are easier to identify clinically, and specific testing for these syndromes is warranted in addition to a microarray. These syndromes include X-linked forms of ID (XLID) in boys and Rett syndrome in girls. In boys, XLID is believed to account for 10% of ID, and approximately 90 XLID genes have been identified to date.7 A family history suggestive of X-linked inheritance is highly predictive of a mutation and indicates the need for specific testing. In girls, MECP2 gene studies are positive in approximately 1.5% of screened individuals with clinical criteria suggestive for Rett syndrome,1 including acquired microcephaly, loss of purposeful hand movements with concomitant development of hand wringing, and loss of verbal and gross motor skills, following a period of at least 6 months of normal development. In each of these cases, specific testing would be an adjunct to (not a replacement for) chromosomal microarray.
Expert consultation and family counseling
The pediatrician will be the first to identify ID/GDD without an identified cause after the history and physical examination. At this point, samples for fragile-X testing and chromosomal microarray may be sent after appropriate pretest counseling. Although most geneticists would welcome consultation at any point in this evaluation, and a pediatrician may choose to defer this process to a genetics consultant, the family may have physical or financial barriers to accessing consultants. Regardless of the clinician who initiates genetic testing, taking this first step provides a significant likelihood of finding a diagnosis. If the results of the array are inconclusive or require further testing and therefore genetics consultation, the family is at least 1 step further along in the process.
Whether a chromosomal array is ordered by the pediatrician or a consultant, the family should be appropriately counseled by the person sending the test. At a minimum, the pediatrician should make the family aware of the possible outcomes of testing. The first possibility is that a diagnosis will be made. If this is the case, management may or may not change, but a diagnosis may predict other organ-system involvement that has not yet manifested, and it may have implications for access to services as well as for health issues in other family members. A second possibility is that a copy-number variation is identified, but it has unknown significance and causality cannot be established. This may require additional testing of the patient and the parents as well as other family members and will require the involvement of specialists. The third possibility is that no abnormality is detected. Other avenues may be pursued, or the patient will have to be reevaluated after a period of time. Finally, there is a chance that the family finds out something unrelated to ID that they do not want to know about, such as deletions or duplications involving a cancer or late-onset neurologic disease. In addition, the parents must be informed that consanguinity will be detected when SNP arrays are used. The significance of this counseling is emphasized by the American Academy of Pediatrics policy statement, “Ethical and Policy Issues in Genetic Testing and Screening of Children,” published in February 2013.17
The importance of establishing a diagnosis cannot be overstated. It can stop the diagnostic odyssey and permits accurate counseling about prognosis and recurrence risk. If a diagnosis is not established after an initial evaluation, the patient should be retested every 6 to 12 months during the first 3 years of life and every 1 to 2 years thereafter. It is worthwhile to initiate a new genetic evaluation in an older patient who may have only been offered karyotyping early in life. This is especially important if reproductive capacity is a concern or if other siblings may be carriers. These patients and their family members are just as likely to benefit from a diagnosis, and there is a much higher probability of diagnosis with chromosomal microarrays.
Other diagnostic procedures
The current data suggest that chromosomal microarrays are a first-tier test in the evaluation of ID and, in addition to fragile-X testing, should be considered second only to an appropriate history, physical examination, and pedigree (Figure 2).3 An additional early diagnostic tool is magnetic resonance imaging (MRI) of the brain. This is specifically warranted when there are focal or diffuse findings on neurologic examination or the presence of macrocephaly, microcephaly, or facial abnormalities suggestive of brain malformation. Additionally, a history of focal or intractable seizures, developmental regression, progressive neurologic deterioration, or movement abnormalities may suggest that an MRI will reveal abnormal structure.3,9 However, it should be noted that abnormal MRI findings may only further characterize a process without necessarily providing an underlying etiology.9
Costs and insurance coverage
Both CGH and SNP arrays are commercially available and provide more sensitive analysis of the genome than a karyotype for a comparable price (the cost of a karyotype is $700 to $1,200; a SNP array costs $1,500 to $2,000). These tests are covered by private insurance companies, Medicaid, and Medicare, but they may require a letter of medical necessity, depending upon the insurer. Whole-exome analysis is the next advance in genetic testing and offers sequencing of about 90% of the protein-coding region of the genome for the same cost as sequencing just a few genes. It will likely increase the sensitivity of detecting genetic changes in ID in the future. This technology is rapidly evolving and the counseling required is quite complex, both before and after testing. Therefore, if microarray and fragile-X testing are unrevealing, a genetics consultation is the next step for the patient with ID.
Summary
Intellectual disability is a prevalent condition that pediatricians should expect to encounter. Establishing a diagnosis is mandatory for optimal patient care, patient and family counseling about prognosis and recurrence risk, and access to health care services. Because of the predominance of genetic causes of ID, chromosomal microarray (CGH or SNP) is a critical second step after a detailed history and physical examination. With sufficient knowledge, pediatricians can begin the process of genetic diagnosis until specialist consultation becomes necessary. A case of ID/GDD of unknown etiology should be considered a diagnosis in progress and should be reevaluated as genetic testing continues to evolve.
References
1. Michelson DJ, Shevell MI, Sherr EH, Moeschler JB, Gropman AL, Ashwal S. Evidence report: genetic and metabolic testing on children with global developmental delay: report of the Quality Standards Subcommittee of the American Academy of Neurology and the Practice Committee of the Child Neurology Society. Neurology. 2011;77(17):1629-1635.
2. Boyle CA, Boulet S, Schieve LA, et al. Trends in the prevalence of developmental disabilities in US children, 1997-2008. Pediatrics. 2011;127(6):1034-1042.
3. Mefford HC, Batshaw ML, Hoffman EP. Genomics, intellectual disability, and autism. N Engl J Med. 2012;366(8):733-743.
4. Miller DT, Adam MP, Aradhya S, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am J Hum Genet. 2010;86(5):749-764.
5. Maulik PK, Mascarenhas MN, Mathers CD, Dua T, Saxena S. Prevalence of intellectual disability: a meta-analysis of population-based studies. Res Dev Disabil. 2011;32(2):419-436. Erratum in: Res Dev Disabil. 2013;34(2):729.
6. Manning M, Hudgins L; Professional Practice and Guidelines Committee. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12(11):742-745.
7. Ropers HH. Genetics of intellectual disability. Curr Opin Genet Dev. 2008;18(3):241-250.
8. Rauch A, Hoyer J, Guth S, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A. 2006;140(19):2063-2074.
9. Moeschler JB. Genetic evaluation of intellectual disabilities. Semin Pediatr Neurol. 2008;15(1):2-9.
10. Schaaf CP, Wiszniewska J, Beaudet AL. Copy number and SNP arrays in clinical diagnostics. Annu Rev Genomics Hum Genet. 2011;12:25-51.
11. Tzetis M, Kitsiou-Tzeli S, Frysira H, Xaidara A, Kanavakis E. The clinical utility of molecular karyotyping using high-resolution array-comparative genomic hybridization. Expert Rev Mol Diagn. 2012;12(5):449-457.
12. Taylor MR, Jirikowic J, Wells C, et al. High prevalence of array comparative genomic hybridization abnormalities in adults with unexplained intellectual disability. Genet Med. 2010;12(1):32-38.
13. Männik K, Parkel S, Palta P, et al. A parallel SNP array study of genomic aberrations associated with mental retardation in patients and general population in Estonia. Eur J Med Genet. 2011;54(2):136-143.
14. Hochstenbach R, van Binsbergen E, Engelen J, et al. Array analysis and karyotyping: workflow consequences based on a retrospective study of 36,325 patients with idiopathic developmental delay in the Netherlands. Eur J Med Genet. 2009;52(4):161-169.
15. Sagoo GS, Butterworth AS, Sanderson S, Shaw-Smith C, Higgins JP, Burton H. Array CGH in patients with learning disability (mental retardation) and congenital anomalies: updated systematic review and meta-analysis of 19 studies and 13,926 subjects. Genet Med. 2009;11(3):139-146.
16. Zahir F, Friedman JM. The impact of array genomic hybridization on mental retardation research: a review of current technologies and their clinical utility. Clin Genet. 2007;72(4):271-287.
17. Committee on Bioethics, Committee on Genetics, American College of Medical Genetics, Genomics Social, Ethical, and Legal Issues Committee. Policy statement: ethical and policy issues in genetic testing and screening of children. Pediatrics. 2013;131(3):620-622.
DR HABELA is a child neurology resident, Department of Neurology, Division of Child Neurology, Johns Hopkins University School of Medicine, Baltimore, Maryland. DR HAMOSH is a professor, Department of Pediatrics and McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore. The authors have nothing to disclose in regard to affiliations with or financial interests in any organizations that may have an interest in any part of this article.